Publications
For the development of accurate and efficient computational methods we combine techniques from combinatorial optimization and machine learning. For instance, we have developed exact and approximate algorithms for a graph coloring problem to increase the resolution of experimental protein structure data, used neural networks to project high-dimensional cellular measurments to an interpretable low-dimensional space, and extended dynamic time warping to the comparison of complex trajectories of, e.g., differentiating immune cells.
The software tools we develop address important biological and medical questions. In close collaborations with biologists and clinicians we have contributed, for example, to the discovery of the embryonic origin of adult neural progenitors, and to linking TIM-3 expression to increased relapse risk in pediatric patients with acute lymphoblastic leukemia.
Highlighted
The link between advances in algorithmic theory and new insights into fundamental problems in biology and human disease is a central characteristic of the research in our lab. The following two publications span the fields of approximation algorithms, wet lab experiments, and applications in neurobiology.
All
2025
2024
2023

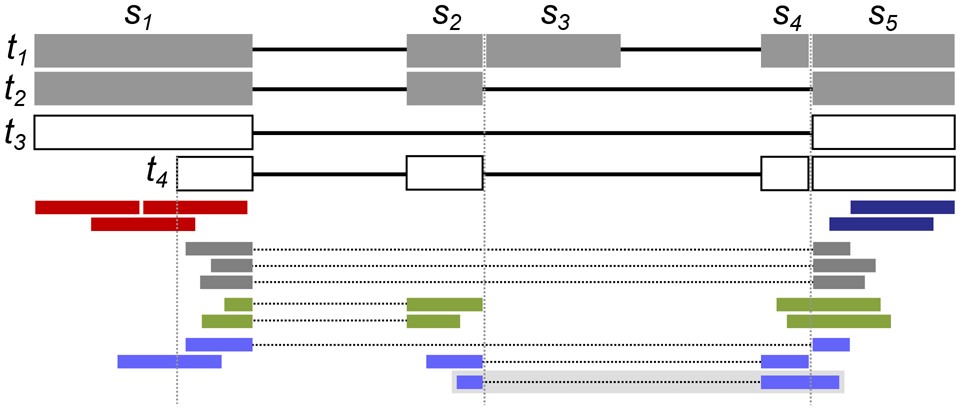
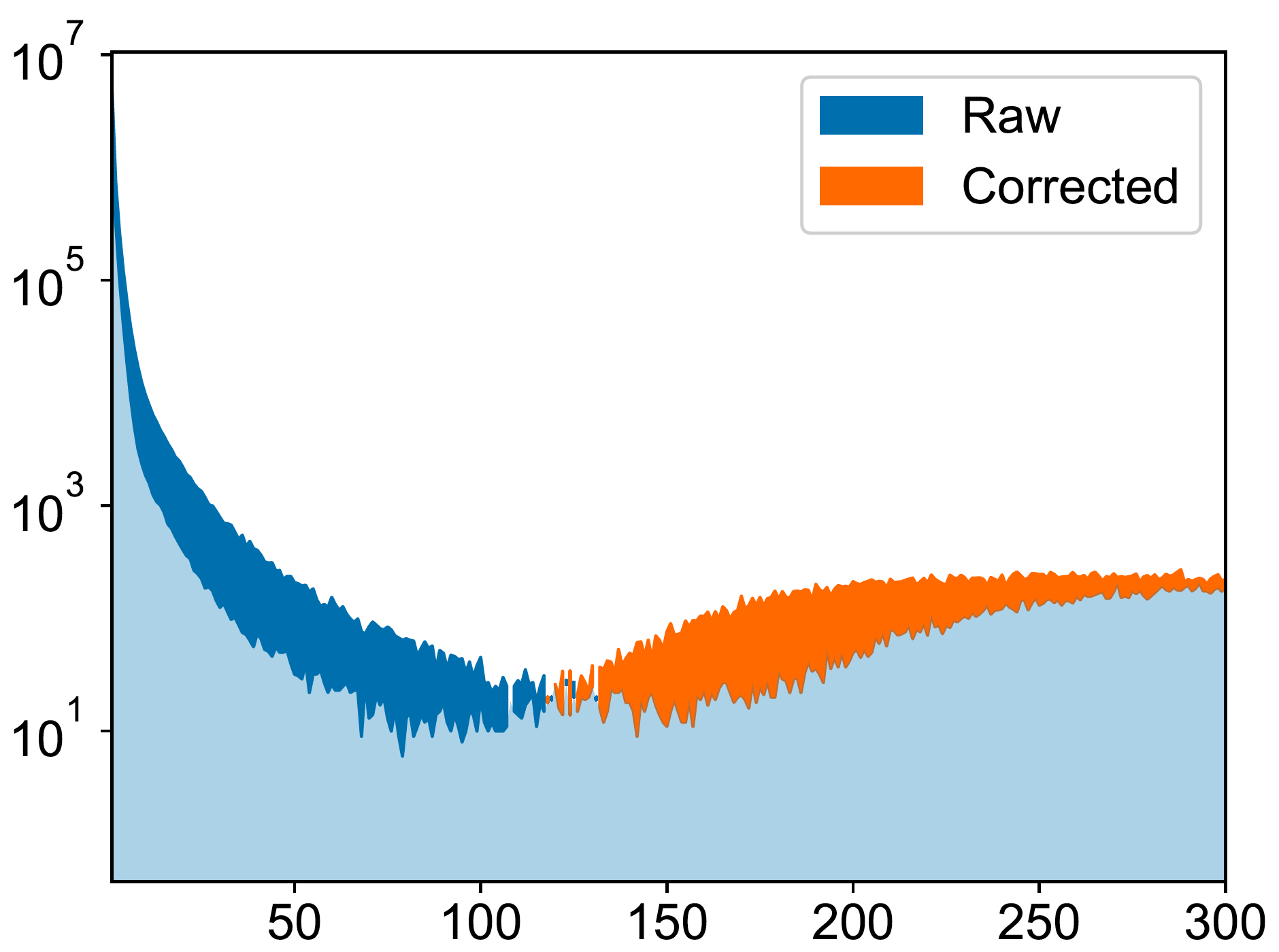
2022
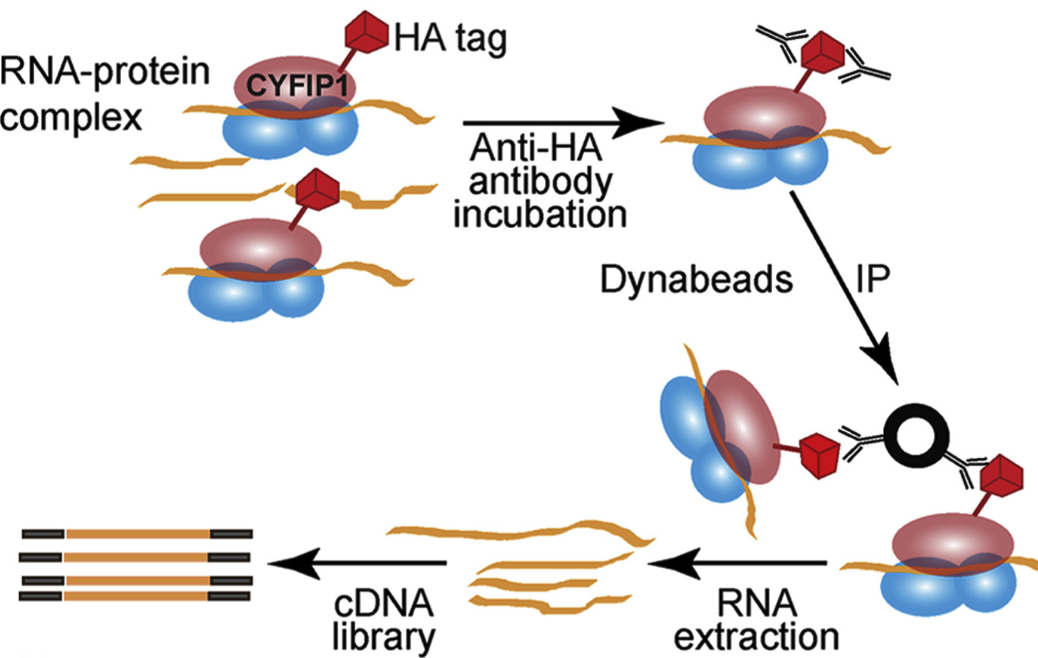
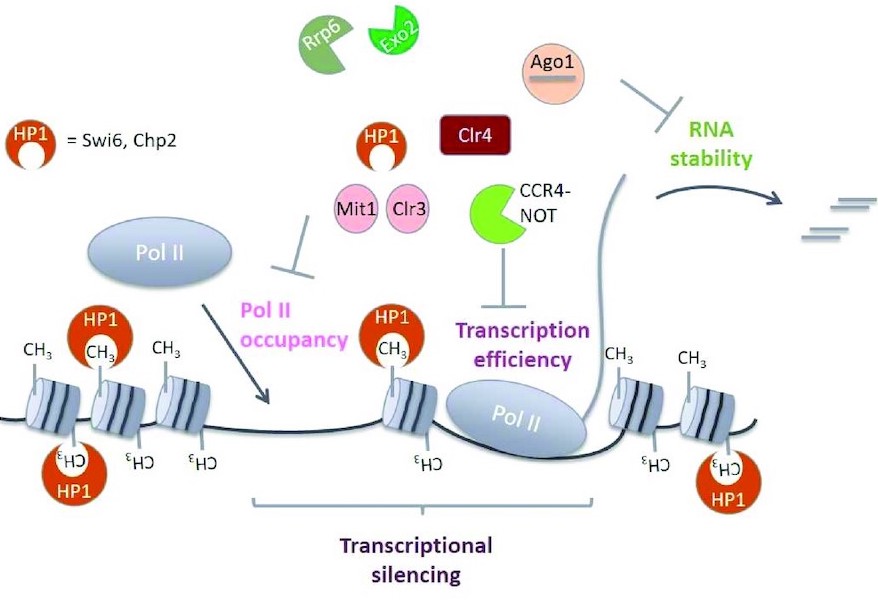
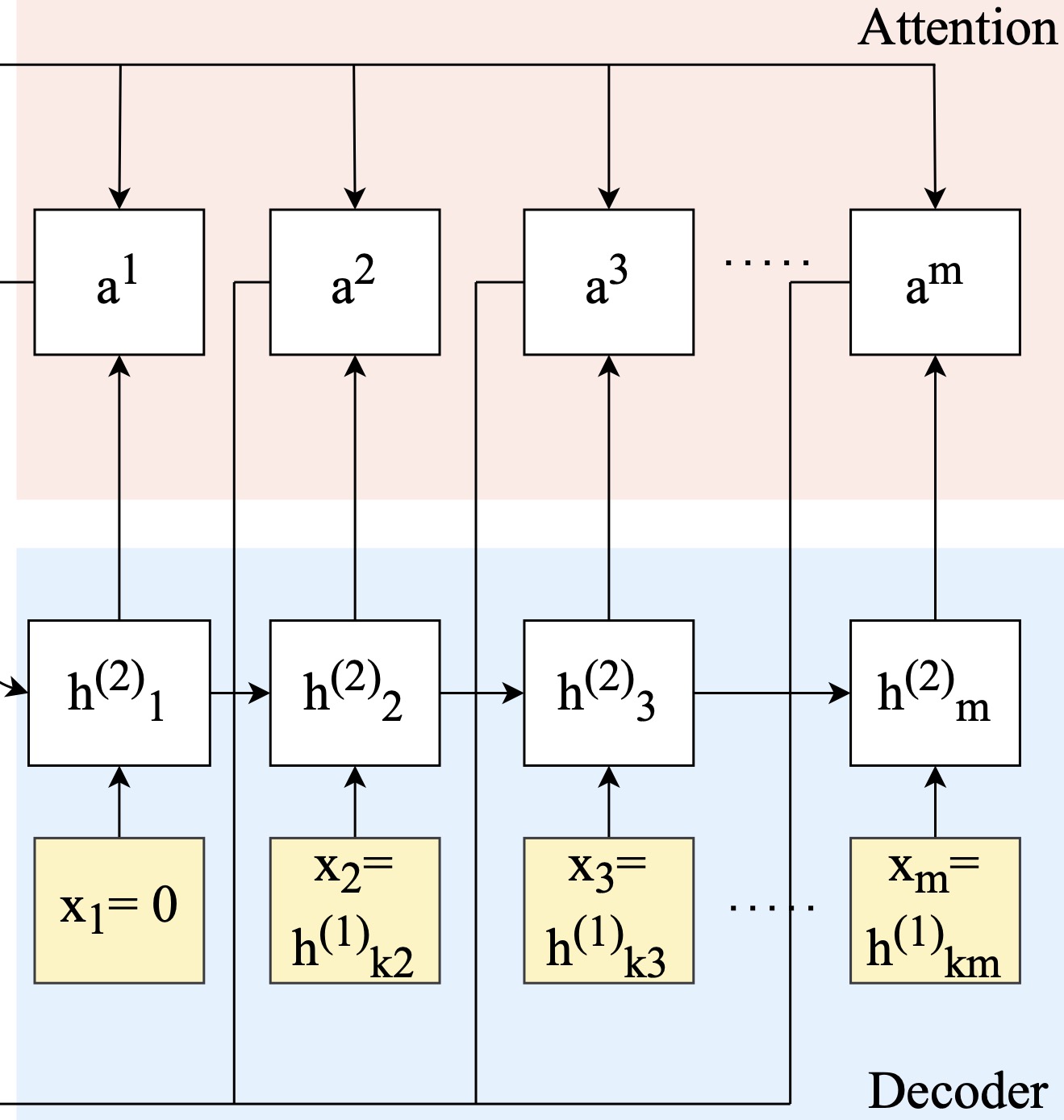

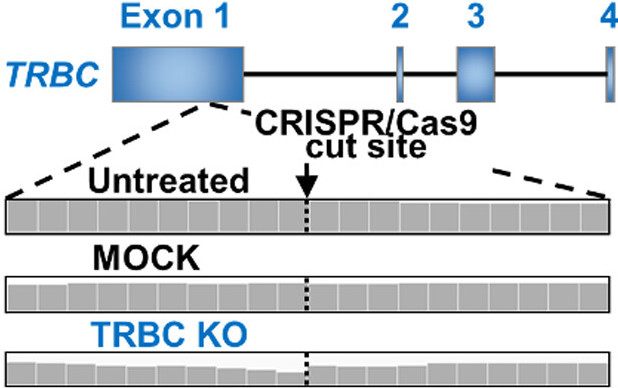
2021


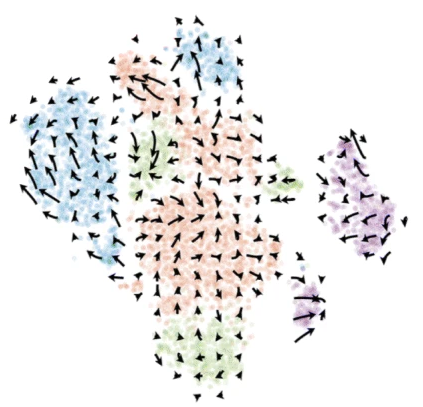

2020








2019
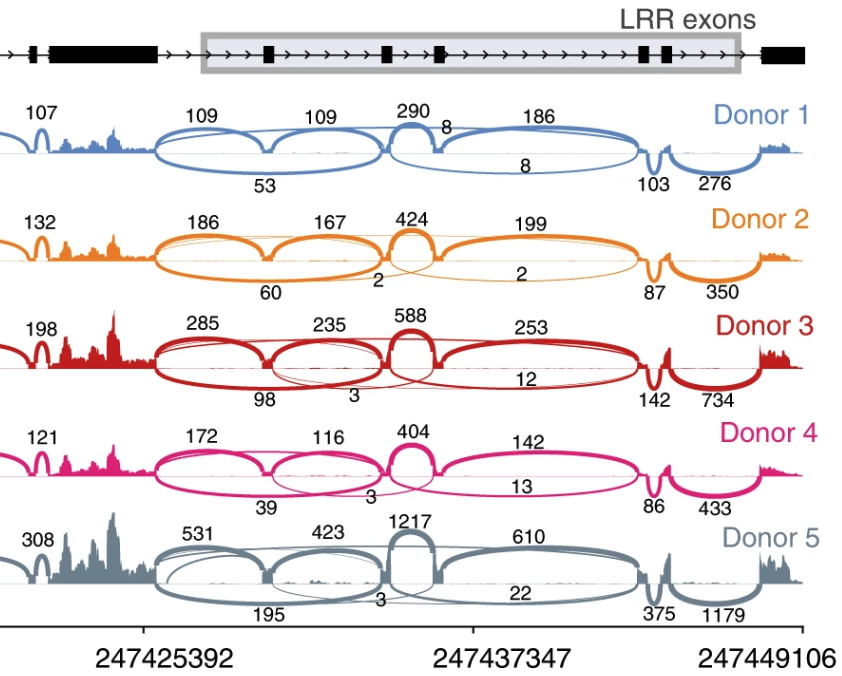





2018





2017


2016







2015

2013

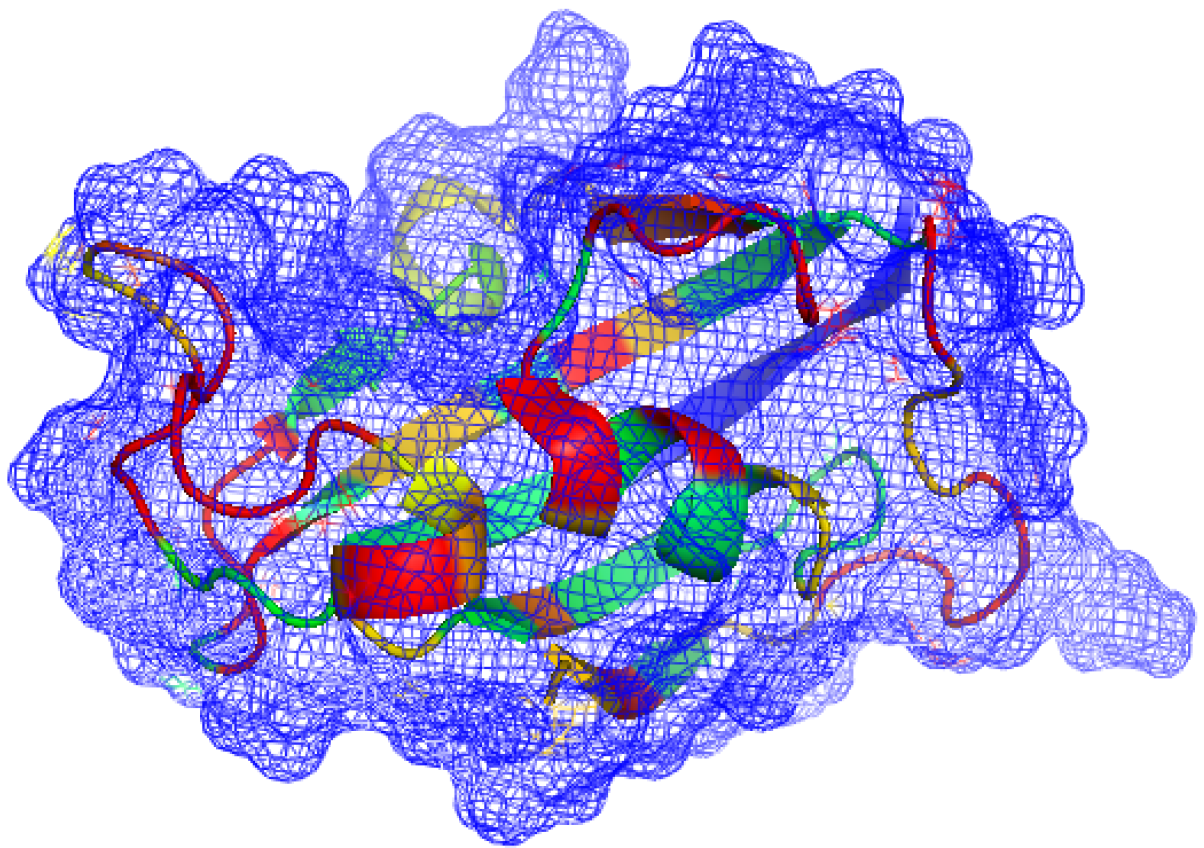




2012



2011



2010




2009

2008



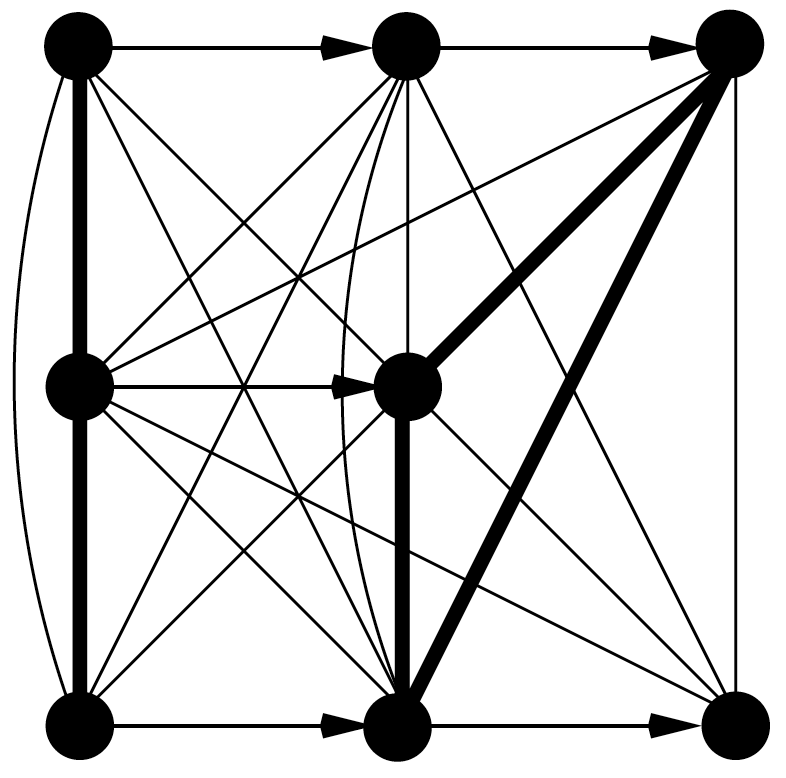
2007

2006

